Linearity of BSA Using Absorbance & Interference Optics
Content Type: Application Note
Dean Clodfelter, M.S., Chad Schwartz, Ph.D., Beckman Coulter Inc., 5350 Lakeview Parkway Dr., Indianapolis, IN 46268
Abstract
The Optima AUC is the latest innovation from Beckman Coulter and offers significant advantages as an analytical ultracentrifugation platform over the predecessor ProteomeLab. Besides the plethora of advancements in user experience on the new platform, the Optima AUC’s optical, thermal, and drive systems have also been upgraded significantly, creating an overall better experience to the end-user in terms of data quality and ease of use. Here, a very well-characterized protein, bovine serum albumin (BSA), was used as the model system to test for linearity of concentration dependence on the oligomerization state.
Introduction
Analytical ultracentrifugation (AUC) characterization of BSA dates back several decades and the highly stable protein is informally used as a reference material to perform instrument and consumable quality control. AUC is a well-regarded technique that dates back to the 1920’s that combines centrifugal force with optical systems to measure the sedimentation of particles over time. Primarily used for characterization of proteins in terms of size, shape, molecular weight, and purity, AUC has become popular in a wide variety of applications including viral vectors, aggregation, nanoparticles, peptides, liposomes, drug conjugates, formulation development and control, and thermodynamic studies. The technique offers significant advantages over other orthogonal biophysical characterization tools in that analysis is performed free in-solution, has limited buffer constraints, and is governed by the first principles of thermodynamics, such that standards are not required.
The ProteomeLab series of AUC instruments, manufactured by Beckman Coulter, were originally designed in the early 1990’s and have been the staple of all AUC analysis over the last two and a half decades. In late 2016, the Optima AUC was launched, and with it, a number of new applications stirred in the minds of researchers globally due to the instrument’s multi wavelength capability. AUC has previously been utilized to assess the purity, safety, and time action of biological therapeutic products, however, the technology is increasingly being used in quality control environments to help assess the overall biological properties of drug products. This has required a greater understanding of the regulatory requirements of validated methods as applied to AUC. The ICH guideline Q2(R1) for Validation of Analytical Procedures includes an assessment of method linearity and method detection and quantitation limits. Serial dilutions of BSA in PBS were evaluated for method linearity and signal to noise as part of this study to evaluate the capabilities of AUC to support method validation according to ICH Q2(R1).
Materials
Albumin from bovine serum (BSA) was purchased from Sigma-Aldrich.
Optima AUC, ProteomeLab XL I, two sector sedimentation velocity analytical cells, quartz windows, An 50 Ti rotor, and torque stand were from Beckman Coulter, Inc.
Methods
BSA Sample Preparation
BSA stock solution was prepared by dissolving BSA powder into PBS to a concentration of 1.5 mg/mL. BSA was then diluted in a 7-fold series dilution with PBS and the optical density of the samples was measured using a Beckman-Coulter DU730 UV/Vis Spectophotometer with a 10mm pathlength at 280 nm.
Instrument Settings
The reference buffer matching the sample solvent was PBS buffer. The 2-sector analytical cells were loaded with 440 μl of both reference and sample buffer. The cells were aligned in the An 50 Ti rotor, and equilibrated at 20˚C for greater than 1 hour. Samples were subsequently spun at 42,000 rpm, 20˚C, 6 hours scanning at Abs280nm and for Rayleigh interference in continuous mode. Acquisition speed for the Optima AUC was set at 20 seconds per cell for absorbance with a 10 μm radial resolution and 20 seconds per cell for interference. The method above was executed on the Beckman Optima AUC and the Beckman ProteomeLab XL-I AUC instruments using the same samples assembled in the same analytical cells shaken after each run.
 |
 |
|
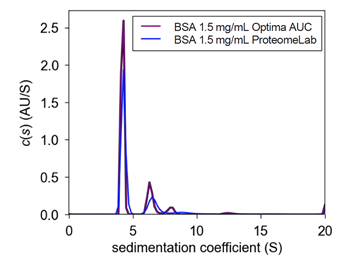
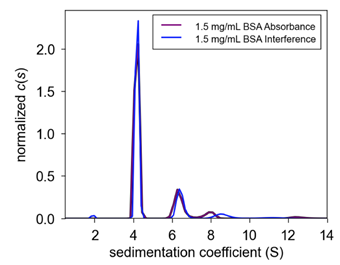
| Figure 1. Comparison c(s) plot of 1.5 mg/mL between ProteomeLab & Optima AUC BSA sedimentation coefficient c(s) plot on the same cell between instruments. The following plot demonstrates the similarity of BSA monomer sedimentation coefficient value but the difference at higher order species. | Figure 2. Interference vs. Absorbance c(s) plot of 1.5 mg/mL Optima AUC BSA sedimentation coefficient c(s) plot on the same cell using different optical systems on the Optima AUC. The following plot demonstrates the similarity in answer between the absorbance and interference optical systems on the Optima AUC. The small differences are most likely due to user load error (meniscus mismatch, inexact buffer mismatch, cell alignment) or cell defects (window scratching, centerpiece defect, residual material). |
Data Analysis†
Data was extracted from the AUC controller and imported into SEDFIT 14.7 g4 (www.analyticalultracentrifugation. com). The data was analyzed using the fitting parameters shown in Figure 1. The data was extracted into GUSSI (http://biophysics.swmed.edu/MBR/software.html) and plotted for c(s) with an s value minimum constraint at 0.3S.
Results & Discussion
BSA is a common albumin that forms multimers of monomers, dimers, trimers, and tetramers, and has been very wellcharacterized by analytical ultracentrifugation, often being used as a test for hardware performance. To determine the performance of the new analytical instrument from Beckman Coulter, this protein was used to check for linearity of response. A series of concentrations were run in replicates on both instruments and analyzed through a commonly used analytical software for sedimentation coefficient. The resulting distribution of sedimentation coefficient was integrated to determine multimer assembly.
In figure 1, a plot of BSA sedimentation coefficient at 1.5 mg/mL tracked using ABS280nm is displayed for both the ProteomeLab and Optima AUC. As expected, BSA will self-associate to four separate species of monomers to tetramers at this concentration which can be accounted for on both instruments. However, as seen in Figure 1, the Optima AUC has a more resolved peak at 12.5 S compared to the ProteomeLab, which is related to the sensitivity of the new instrument. As the light intensity has previously been characterized to be very similar on both instruments, the increased sensitivity is correlated to less systematic noise.
The Rayleigh interference system was also used in each replicate experiment and a plot of the sedimentation coefficient as characterized by both optical systems is shown in Figure 2. Here, it is observed that both optical systems generate a similar answer.
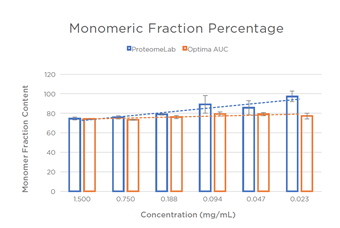
In order to better understand the increased sensitivity and precision of the Optima AUC, a number of analyses were performed on the replicated experiments for each instrument. In Figure 3, the monomer response is plotted among concentrations and analyzed for linearity of response. The slope of monomer response for the ProteomeLab trends upward as the concentration is decreased, suggesting the fraction of monomer increases as concentrations decrease. The same slope for the Optima AUC is nearly flat, suggesting a repeatable monomeric response (~78% monomer at all concentrations). Since we know that proteins disassemble at lower concentrations, it is intriguing that the portion of monomeric content remains stable within these working concentrations and conditions. It is hypothesized that the higher signal to noise ratio of the Optima AUC versus the ProteomeLab (and other analytical equipment) allows the instrument to better detect the higher order species. It should be noted that the cell holding 0.375 mg/mL BSA leaked and was not analyzed.
 |
 |
|
| Figure 3: Linearity of Monomeric Fraction Percentage Percentage of monomeric species at 6 concentrations of BSA. |
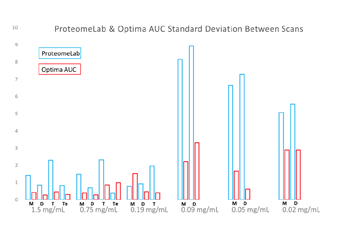
Figure 4. Standard Deviation of Dilution Series for Each Oligomeric State Standard deviation as a percentage of species among replicates at each concentration for stoichiometry. |
Additionally, it was observed that the variability in response was greater in the ProteomeLab than the Optima AUC. The standard deviation of each replicated experiment for each oligomer was plotted in Figure 4. In general, standard deviation of response is much larger for the ProteomeLab than the Optima AUC, suggesting higher repeatability with the new instrument.
Finally, to expand on the results for linearity of response in Figure 3, a separate analysis was performed for each multimer on both instruments. In Figure 5, these values are plotted. At low concentrations, there is significant difference in the apparent association states between the Optima AUC and the ProteomeLab. The apparent percentage of lower-order species (monomers, dimers) is significantly higher in the ProteomeLab than the Optima AUC. Again, it is suggested that the Optima AUC has a lower detection and quantitation limits when compared to the ProteomeLab. Additionally, on the plot, it is clear that percent monomer decreases, but percent dimer and trimer increases linearly as concentration increases for the ProteomeLab. However, the multimeric response on the Optima AUC is a flat line, suggesting repeatability of response and higher sensitivity at all concentrations for each multimeric state.
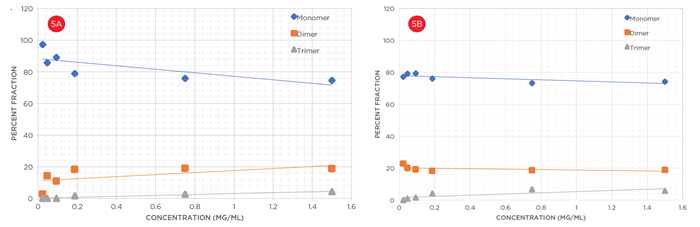 |
| Figure 5: Linearity of Combined Stoichiometry Apparent self-association of BSA over a 6-fold dilution series between (a) ProteomeLab and (b) Optima AUC. |
CENT-2713APP05.17

